

Figure: Digital pathology for image analysis.
Histopathology, the microscopic analysis of tissue sections, serves as a cornerstone for diagnosing and characterizing various diseases, particularly cancer [2,3]. The traditional diagnostic workflow involves the manual examination of Hematoxylin and Eosin (H&E)-stained slides by expert pathologists, relying on morphological patterns to guide clinical decisions. Despite its reliability, this manual process is time-intensive, subject to intra- and inter-observer variability, and increasingly burdensome given the rising volume of clinical cases. The emergence of deep learning methods has significantly advanced the field of computational histopathology, allowing for high-throughput and scalable analysis of gigapixel whole slide images (WSIs) [4]. These digital scans, however, pose substantial computational challenges due to their massive resolution and complex tissue structures. Multiple Instance Learning (MIL) has gained popularity for slide-level classification by dividing WSIs into smaller, manageable patches. Yet, standard MIL assumes independence among instances, ignoring spatial and contextual dependencies that are crucial for accurate histopathological assessment [6].
Microscopic imaging, while more memory-efficient and cost-effective than WSI scanning, introduces additional complications. Unlike scanned WSIs, microscope-based patches often lack absolute spatial coordinates and contain redundancy due to subjective sampling by the pathologist. These factors hinder the construction of structured inputs for downstream learning, especially in models like ABMIL [6], TransMIL [7], and graph-based MIL approaches [8]. Caption generation from histopathological images presents its own set of challenges. Models relying on Recurrent Neural Networks (RNNs), Long Short-Term Memory (LSTM), or spatial feature grids [10] fail to adequately model the complex and dispersed regions of interest typical in pathology slides [9,11].
The ability to automatically generate descriptive captions from WSIs holds immense potential for augmenting diagnostic workflows. Captioning models can help synthesize the visual complexity of pathology images into interpretable textual information, allowing pathologists to quickly identify regions of interest and gain additional context for diagnosis [2,12]. However, the field remains underexplored due to the lack of large, annotated datasets and the inherent complexity of medical images [13]. The growing success of AI-driven diagnostic tools in classification tasks has paved the way for more advanced applications, such as multimodal captioning and interpretability [14,15]. Large Language Models (LLMs), such as LLaMA [16], ClinicalT5 [17], and BioGPT [18], have demonstrated impressive performance in understanding and generating domain-specific text. When combined with Vision Transformers (ViTs) [20], which excel in capturing spatial hierarchies from visual data, these models enable robust multimodal learning pipelines. Such integration facilitates accurate and context-rich caption generation for complex biomedical tasks [21–22]. The proposed research is motivated by this potential and aims to develop hybrid models that effectively leverage both visual and linguistic representations to improve pathological image interpretation and reporting.
The TransUAAE-CapGen model comprises a UNet-based Adversarial Autoencoder (AAE) that serves as a feature extractor. It captures both local and global histopathological features through a deep hierarchical architecture. These features are subsequently passed to a Transformer network designed to generate descriptive captions. The Transformer utilizes self-attention mechanisms and positional encodings to effectively model sequential dependencies, producing accurate and clinically relevant textual descriptions.
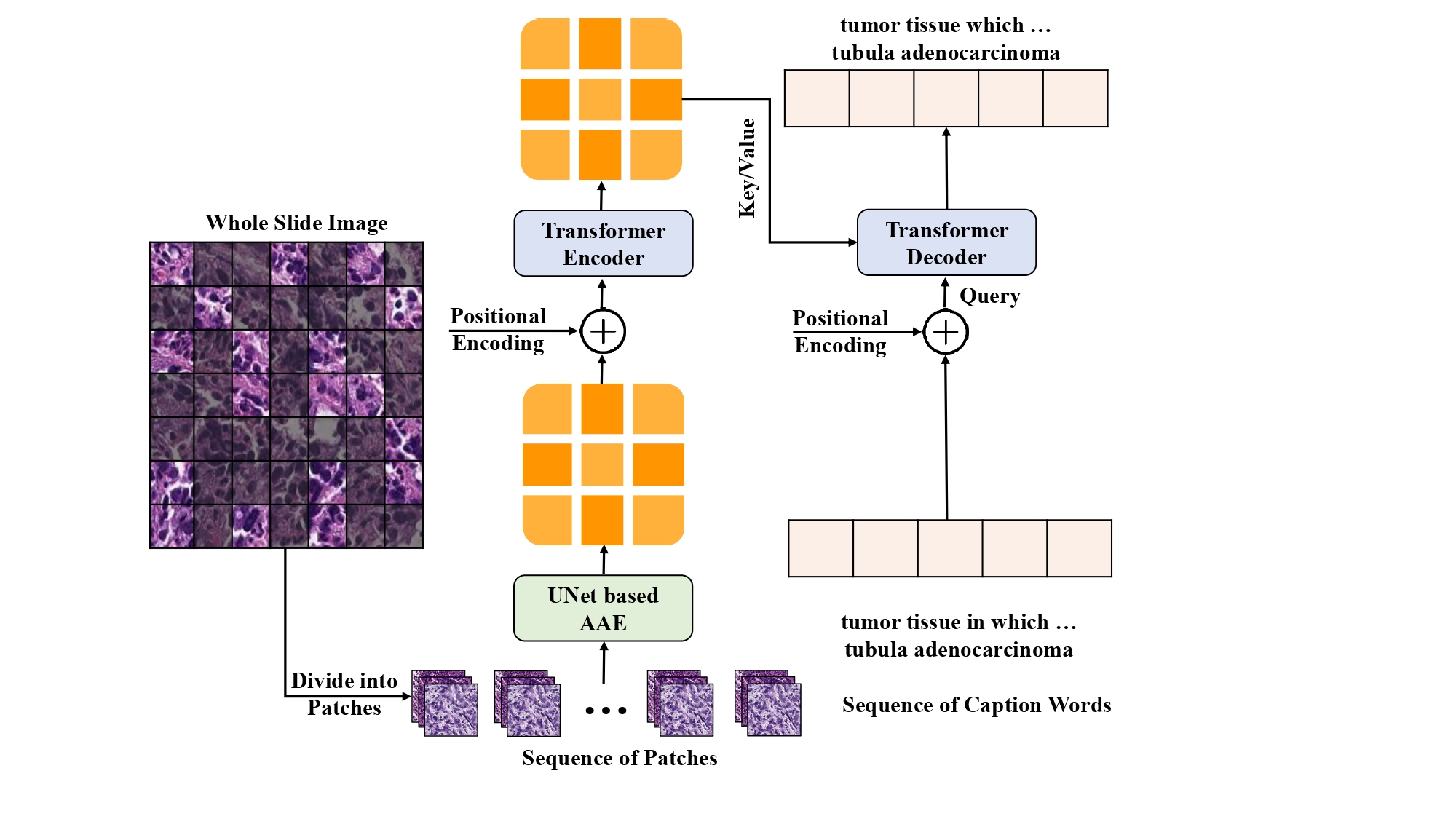
Figure: Architecture of our proposed TransUAAE-CapGen model for histopathological caption generation.
The GNN-ViTCap architecture integrates advanced modules to overcome redundancy and spatial ambiguity. Initially, histopathological patches undergo feature extraction using a Vision Transformer (ViT), converting image patches into meaningful embeddings. An attention-based deep embedded clustering technique then identifies and selects representative patches, reducing data redundancy. Next, Graph Neural Networks (GNN) are employed to construct relational graphs capturing spatial and contextual information between selected representative patches. Lastly, the aggregated embeddings from the GNN are projected into a language model space, integrating with large language models (LLMs) to perform classification and sophisticated caption generation tasks.
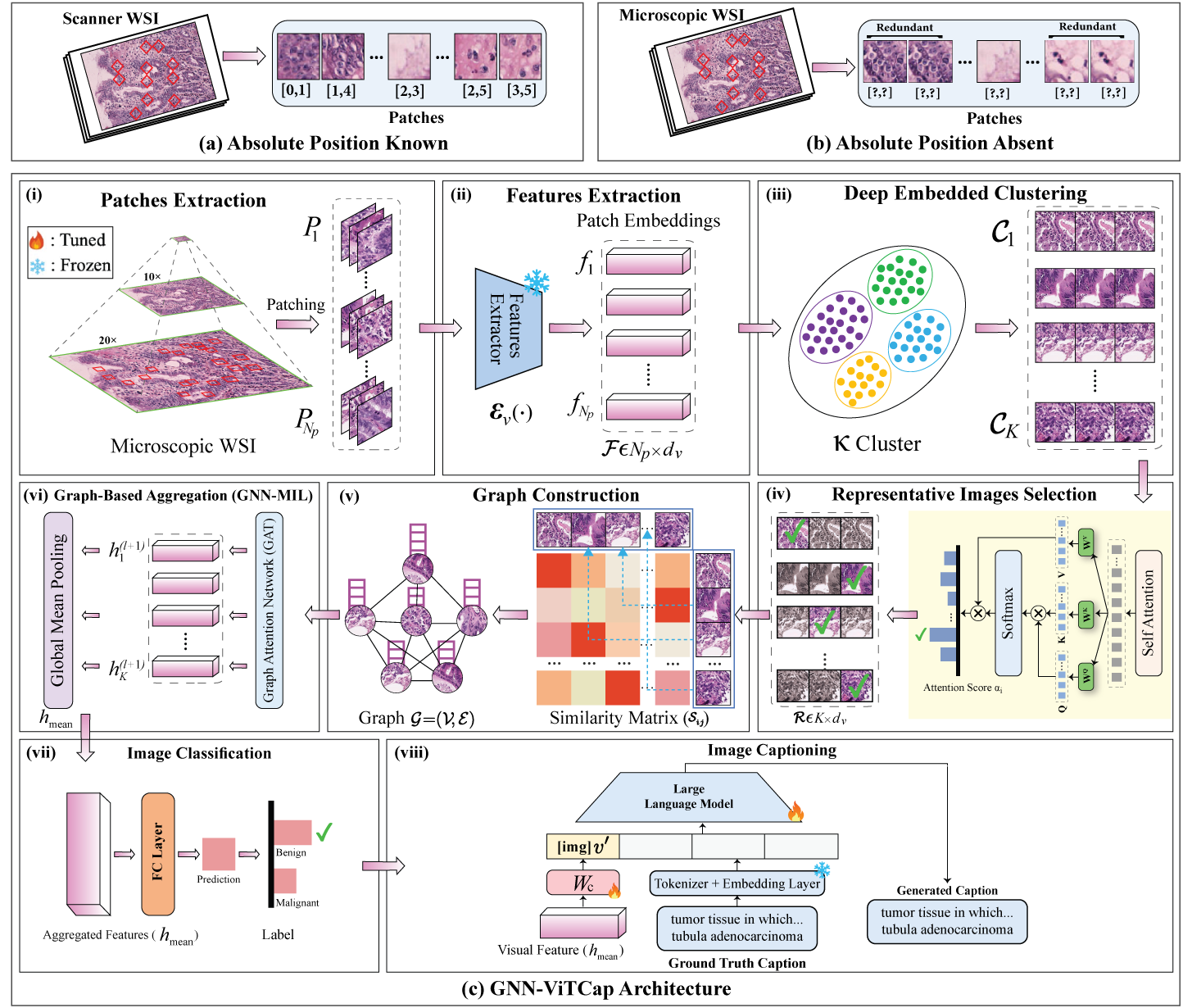
Figure: Overview of the GNN-ViTCap framework for microscopic whole slide images classification and captioning. (a) Scanner WSI where the absolute position of each patch is known. (b) Microscopic WSI lacks patch position and contains redundant patches due to multiple captures from a pathologist’s subjective perspective. (c) GNN-ViTCap architecture: (i) extracting the patches from whole slide image, (ii) extracting image embeddings using a visual feature extractor, (iii) removing redundancy through deep embedded clustering, (iv) extracting representative images with scalar dot attention mechanism, (v) constructing a graph neural network (GNN) using the similarity of representative patches to capture contextual information within clusters (local) and between different clusters (regional), (vi) applying global mean pooling, which aggregates all node representations, (vii) classifying microscopic WSI using aggregated image embeddings and, (viii) projecting the aggregated image embeddings into the language model’s input space using a linear layer, and combining these projections with input caption tokens fine-tunes the LLMs for caption generation.